6. The annotate module¶
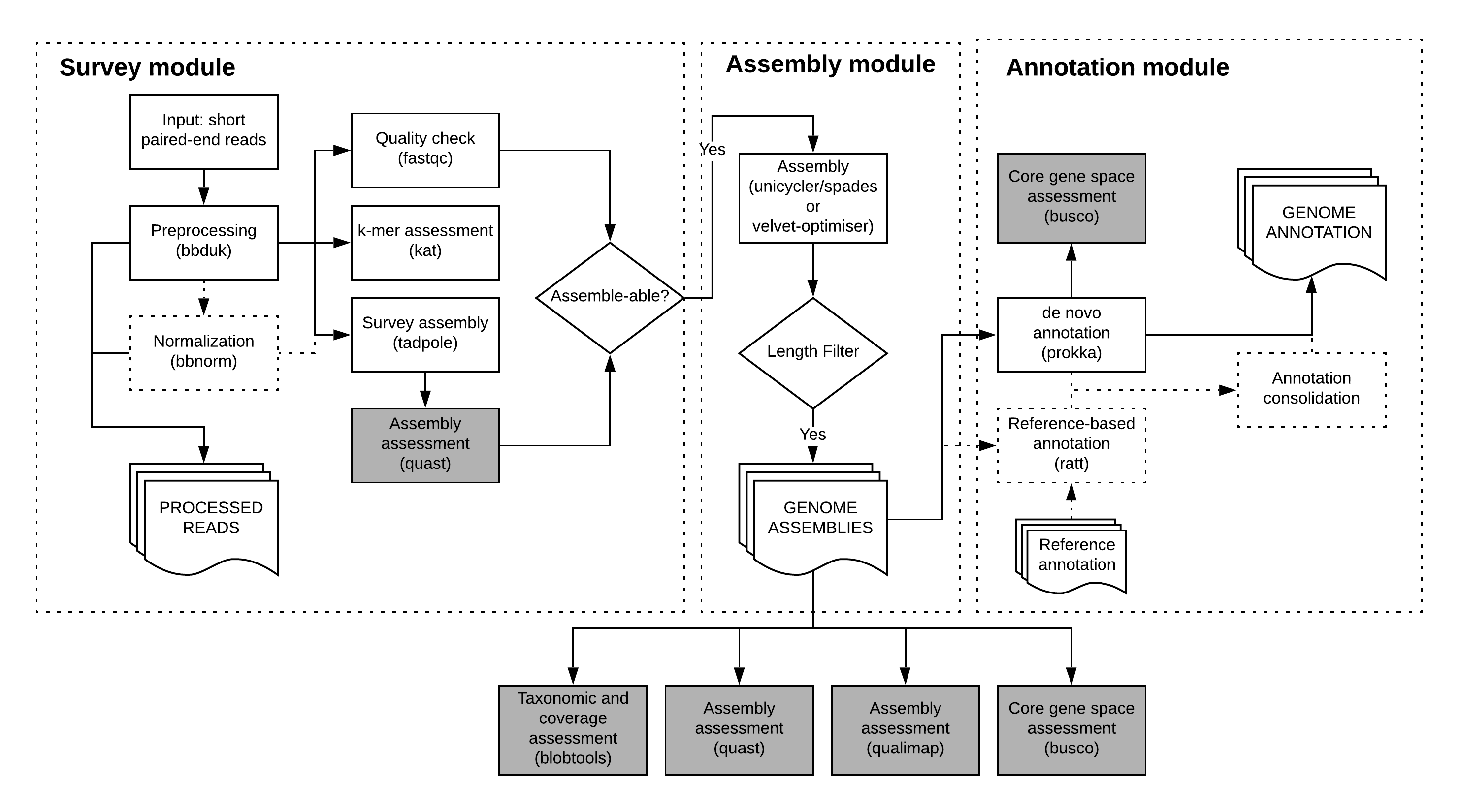
The annotate performs de novo, and optionally reference-based, genome annotation on a set of assemblies.
6.1. Sample sheet preparation¶
If the user has previously run the assemble module without annotation, the resulting samplesheet samplesheet.asm_pass.tsv can be used directly
to drive the annotate module.
Otherwise, the user should prepare a comma-separated samplesheet following the column order below.
- Sample ID
- Sample Name (can be the same as Sample ID; the intention is to allow a more understandable sample reference in the future)
- Full path to assembly file
TODO: CHECK
6.2. Command line arguments¶
The command bgrrl -h or bgrrl <stage> -h (or --help instead of -h) will display a list of command line options.
Remember each bgrr| run requires at the very least the following three command line parameters:
input_sheet--config--hpc_config
6.2.1. annotate options:¶
--custom-prokka-proteins CUSTOM_PROKKA_PROTEINSIf you have a custom protein database that you would like prokka to use (
prokka’s--proteinsoption), then specify the path to it here. [n/a]--ratt-reference RATT_REFERENCEPath to reference data for ratt annotation transfer
--no-packagingDisable automatic packaging. [False]
--prokka-package-style {by_sample,all_in_one}Should the prokka annotation be packaged into one directory per sample (by_sample) or into one single directory (all_in_one)? [by_sample]